ARCensReg R
packageThe ARCensReg package fits a univariate censored linear
regression model with autoregressive (AR) errors. The discrete-time
representation of this model for the observed response at time
t is given by
Yt = xt⊤β + ξt,
ξt = ϕ1ξt − 1 + ϕ2ξt − 2 + … + ϕpξt − p + ηt, t = 1, …, p, p + 1, …, n,
where Yt is the response variable, β is a vector of regression parameters of dimension l, xt is a vector of non-stochastic regressor variables, and ξt is the AR error with ηt representing the innovations and ϕ denoting the vector of AR coefficients. For the innovations ηt, we consider the normal or the Student-t distribution. The maximum likelihood estimates are obtained through the Stochastic Approximation Expectation-Maximization (SAEM) algorithm (Delyon, Lavielle, and Moulines 1999), while the standard errors of the parameters are approximated by the Louis method (Louis 1982). This package also predicts future observations and supports missing values on the dependent variable. See, for instance, (Schumacher, Lachos, and Dey 2017) and (Valeriano et al. 2021).
For the normal model, influence diagnostic could be performed by a local influence approach (Cook 1986) with three possible perturbation schemes: response perturbation, scale matrix perturbation, or explanatory variable perturbation. For more details see (Schumacher et al. 2018).
The ARCensReg package provides the following
functions:
ARCensReg: fits a univariate censored linear regression
model with autoregressive errors under the normal distribution.ARtCensReg: fits a univariate censored linear
regression model with autoregressive errors considering Student-t
innovations.InfDiag: performs influence diagnostic by a local
influence approach with three possible perturbation schemes.rARCens: simulates a censored response variable with
autoregressive errors of order p.residuals: computes conditional and quantile
residuals.predict, print, summary, and
plot functions also work for objects given as an output of
functions ARCensReg and ARtCensReg. Function
plot also has methods for outputs of functions
InfDiag and residuals.
Next, we will describe how to install the package and use all the previous methods in artificial examples.
The released version of ARCensReg from CRAN can be installed with:
install.packages("ARCensReg")And the development version from GitHub with:
# install.packages("devtools")
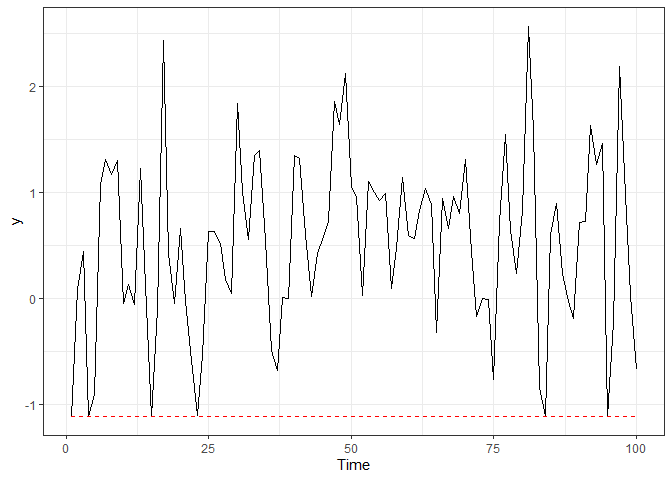
devtools::install_github("fernandalschumacher/ARCensReg")Example 1. We simulated a dataset of length n = 100 from the autoregressive model of order p = 2 with normal innovations and left censoring.
library(ARCensReg)
library(ggplot2)
set.seed(12341)
n = 100
x = cbind(1, runif(n))
dat = rARCens(n=n, beta=c(1,-1), phi=c(.48,-0.4), sig2=.5, x=x,
cens='left', pcens=.05, innov="norm")
ggplot(dat$data, aes(x=1:n, y=y)) + geom_line() + labs(x="Time") + theme_bw() +
geom_line(aes(x=1:n, y=ucl), color="red", linetype="dashed")
Supposing the AR order is unknown, we fit a censored linear regression model with Gaussian AR errors of order p = 1 and p = 2, and the information criteria are compared.
fit1 = ARCensReg(dat$data$cc, dat$data$lcl, dat$data$ucl, dat$data$y, x,
p=1, pc=0.15, show_se=FALSE, quiet=TRUE)
fit1$critFin
#> Loglik AIC BIC AICcorr
#> Value -113.027 234.054 244.475 234.475
fit2 = ARCensReg(dat$data$cc, dat$data$lcl, dat$data$ucl, dat$data$y, x,
p=2, pc=0.15, quiet=TRUE)
fit2$critFin
#> Loglik AIC BIC AICcorr
#> Value -105.279 220.558 233.583 221.196Based on the information criteria AIC and BIC, the model with AR
errors of order p = 2 is the best fit for this data. The
parameter estimates and standard errors can be visualized through
functions summary and print.
summary(fit2)
#> ---------------------------------------------------
#> Censored Linear Regression Model with AR Errors
#> ---------------------------------------------------
#> Call:
#> ARCensReg(cc = dat$data$cc, lcl = dat$data$lcl, ucl = dat$data$ucl,
#> y = dat$data$y, x = x, p = 2, pc = 0.15, quiet = TRUE)
#>
#> Estimated parameters:
#> beta0 beta1 sigma2 phi1 phi2
#> 1.0830 -1.0965 0.4713 0.4690 -0.3883
#> s.e. 0.1465 0.2389 0.0689 0.0949 0.0941
#>
#> Model selection criteria:
#> Loglik AIC BIC AICcorr
#> Value -105.279 220.558 233.583 221.196
#>
#> Details:
#> Type of censoring: left
#> Number of missing values: 0
#> Convergence reached?:
#> Iterations: 168 / 400
#> MC sample: 10
#> Cut point: 0.15
#> Processing time: 44.76831 secsMoreover, for censored data, the convergence plot of the parameter
estimates can be displayed through function plot.
plot(fit2)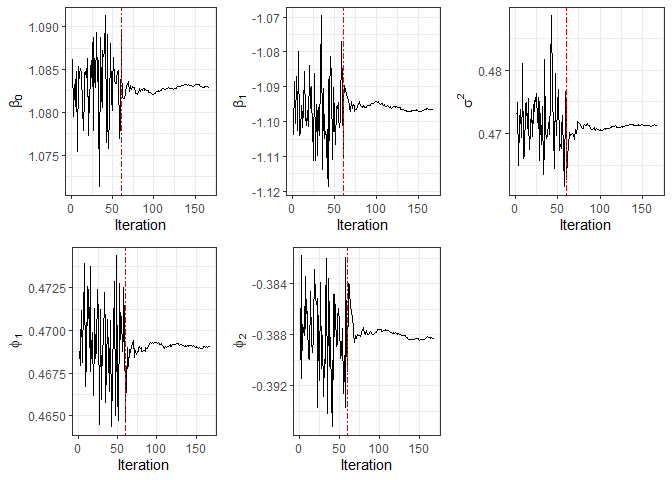
Now, we perturb the observation 81 by making it equal to 6 and then fit a censored linear regression model with Gaussian AR errors of order p = 2.
y2 = dat$data$y
y2[81] = 6
fit3 = ARCensReg(dat$data$cc, dat$data$lcl, dat$data$ucl, y2, x, p=2,
show_se=FALSE, quiet=TRUE)
fit3$tab
#> beta0 beta1 sigma2 phi1 phi2
#> 1.1702 -1.2065 0.7237 0.3812 -0.3355It is worth noting that the parameter estimates were affected because of the perturbation. Thence, we can perform influence diagnostic to identify influential points which may cause unwanted effects on estimation and goodness of fit. In the following analysis, we only consider the response perturbation scheme, where we deduced that observations 80 to 82 may be influential.
M0y = InfDiag(fit3, k=3.5, perturbation="y")
#> Perturbation scheme: y
#> Benchmark: 0.059
#> Detected points: 80 81 82
plot(M0y)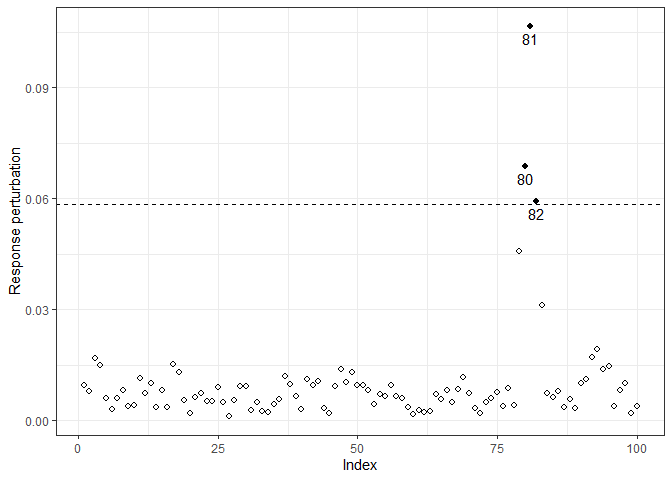
Example 2. A dataset of size n = 200 is
simulated from the censored regression model with Student-t innovations
and right censoring. To fit this data, we can use the function
ARtCensReg, but it is worth mentioning that this function
only works for response vectors with the first p values wholly
observed.
set.seed(783796)
n = 200
x = cbind(1, runif(n))
dat2 = rARCens(n=n, beta=c(2,1), phi=c(.48,-.2), sig2=.5, x=x, cens='right',
pcens=.05, innov='t', nu=3)
head(dat2$data)
#> y cc lcl ucl
#> 1 3.3242078 0 4.284143 Inf
#> 2 2.9409784 0 4.284143 Inf
#> 3 3.5386525 0 4.284143 Inf
#> 4 3.4107191 0 4.284143 Inf
#> 5 0.3564079 0 4.284143 Inf
#> 6 2.6332103 0 4.284143 InfFor models with Student-t innovations, the degrees of freedom can be
provided through argument nufix when it is known, or the
algorithm will estimate it when it is not provided, i.e.,
nufix=NULL.
# Fitting the model with nu known
fit1 = ARtCensReg(dat2$data$cc, dat2$data$lcl, dat2$data$ucl, dat2$data$y, x,
p=2, M=20, nufix=3, quiet=TRUE)
fit1$tab
#> beta0 beta1 sigma2 phi1 phi2
#> 1.9707 0.9594 0.4468 0.3778 -0.1562
#> s.e. 0.1230 0.1890 0.0640 0.0572 0.0590
# Fitting the model with nu unknown
fit2 = ARtCensReg(dat2$data$cc, dat2$data$lcl, dat2$data$ucl, dat2$data$y, x,
p=2, M=20, quiet=TRUE)
fit2$tab
#> beta0 beta1 sigma2 phi1 phi2 nu
#> 1.9630 0.9720 0.4812 0.3798 -0.1563 3.4760
#> s.e. 0.1266 0.1953 0.0991 0.0592 0.0606 1.1295Note that the parameter estimates obtained from both models are close, and the estimate of ν was close to the true value (ν = 3).
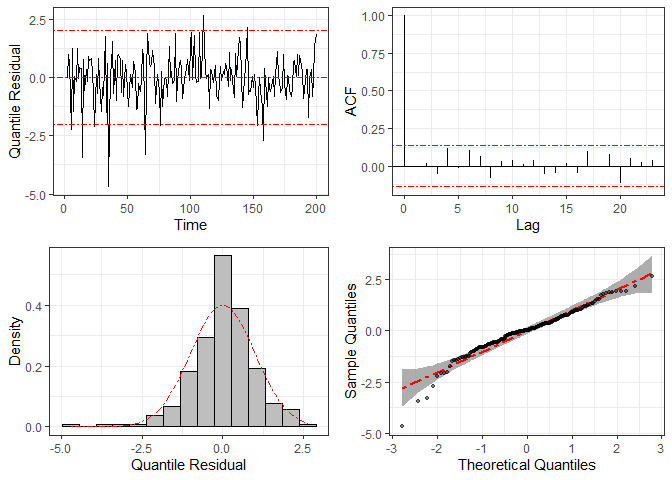
To check the statistical model’s specification, we can use graphical
methods based on the quantile residuals, which are computed through
function residuals and plotted by function
plot.
res = residuals(fit2)
plot(res)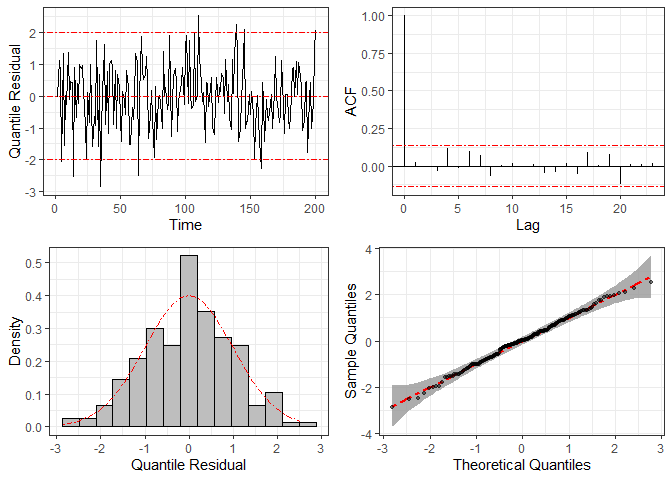
For comparison, we fit the same dataset considering the normal distribution (i.e., disregarding the heavy tails) and compute the corresponding quantile residuals. The resulting plots are given below, where we can see clear signs of non-normality, such as large residuals and some points outside the confidence band in the Q-Q plots.
fit3 = ARCensReg(dat2$data$cc, dat2$data$lcl, dat2$data$ucl, dat2$data$y, x,
p=2, M=20, show_se=FALSE, quiet=TRUE)
plot(residuals(fit3))