

The goal of ViroReportR is to provide a toolbox to
conveniently generate short-term forecasts (with accompanied
diagnostics) for viral respiratory diseases.
ViroReportR depends on the latest version of the
EpiEstim package (2.4). Thus, this version of the package
must be installed from GitHub prior to installing the
ViroReportR package using:
# install.packages("devtools")
install.packages('EpiEstim', repos = c('https://mrc-ide.r-universe.dev', 'https://cloud.r-project.org'))You can then install the development version of
ViroReportR from GitHub
with:
devtools::install_github("BCCDC-PHSA/ViroReportR")ViroReportR can be used to generate short-term forecasts
with accompanied diagnostics in a few lines of code. We go through an
example here where the EpiEstim backend is used to generate
forecasts of Influenza-A, RSV and SARS-CoV-2.
library(ViroReportR)
library(tidyverse)
#> ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
#> ✔ dplyr 1.1.4 ✔ readr 2.1.5
#> ✔ forcats 1.0.0 ✔ stringr 1.6.0
#> ✔ ggplot2 4.0.0 ✔ tibble 3.2.1
#> ✔ lubridate 1.9.4 ✔ tidyr 1.3.1
#> ✔ purrr 1.0.2
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
library(ggplot2)
library(here)
#> here() starts at C:/Users/rebeca.falcao/ViroReportR
library(DT)
library(purrr)
library(kableExtra)
#>
#> Attaching package: 'kableExtra'
#>
#> The following object is masked from 'package:dplyr':
#>
#> group_rowsWe will use simulate_data to simulate date for Influenza
A, RSV, and SARS-CoV-2, which is included with the
ViroReportR package. We then pivot the simulated data to
transform into a dataset with three columns: date,
disease_type and confirm in accordance to
format accepted by the model fitting functions.
diseases<-c("flu_a", "rsv", "sars_cov2")
data <- simulate_data(days=365, #days spanning simulation
peaks = c("flu_a"=90,"rsv"=110,"sars_cov2"=160), #peak day for each disease
amplitudes=c("flu_a"=50,"rsv"=40,"sars_cov2"=20), #amplitude of peak for each disease
scales = c("flu_a"=-0.004,"rsv"=-0.005,"sars_cov2"=-0.001), # spread of peak for each disease
time_offset = 0, #number of days to offset start of simulation
noise_sd = 5, #noise term
start_date = "2024-01-07" #starting day (Sunday)
)
data$date <- lubridate::ymd(data$date)
vri_data_list <- map2(rep(list(data), length(diseases)),
diseases,~ get_aggregated_data(.x, "date", .y)) %>% set_names(diseases)
head(vri_data_list)
#> $flu_a
#> # A tibble: 366 × 2
#> date confirm
#> <date> <dbl>
#> 1 2024-01-07 0
#> 2 2024-01-08 8
#> 3 2024-01-09 0
#> 4 2024-01-10 5
#> 5 2024-01-11 0
#> 6 2024-01-12 8
#> 7 2024-01-13 9
#> 8 2024-01-14 0
#> 9 2024-01-15 4
#> 10 2024-01-16 0
#> # ℹ 356 more rows
#>
#> $rsv
#> # A tibble: 366 × 2
#> date confirm
#> <date> <dbl>
#> 1 2024-01-07 0
#> 2 2024-01-08 0
#> 3 2024-01-09 0
#> 4 2024-01-10 0
#> 5 2024-01-11 5
#> 6 2024-01-12 0
#> 7 2024-01-13 0
#> 8 2024-01-14 1
#> 9 2024-01-15 0
#> 10 2024-01-16 0
#> # ℹ 356 more rows
#>
#> $sars_cov2
#> # A tibble: 366 × 2
#> date confirm
#> <date> <dbl>
#> 1 2024-01-07 9
#> 2 2024-01-08 0
#> 3 2024-01-09 0
#> 4 2024-01-10 0
#> 5 2024-01-11 0
#> 6 2024-01-12 0
#> 7 2024-01-13 0
#> 8 2024-01-14 3
#> 9 2024-01-15 0
#> 10 2024-01-16 0
#> # ℹ 356 more rowsThe code below estimates the reproduction number using
EpiEstim through the generate_forecast()
function for each disease type via the purrr::map2()
function. The generate_forecast() function prepares the
data, estimates the reproduction number, and produces short-term
forecasts of daily confirmed cases for an n_days forecast
horizon. The other current choice for the forecasting algorithm is
EpiFilter (WIP).
# parameters set-up
start_date <- min(data$date) + 13
n_days <- 14 # number of days ahead to forecast (n_days)
smooth <- FALSE # logical indicating whether smoothing should be applied in the forecastforecasts_results <- tibble(
vri_data_list,
forecasts = map2(
vri_data_list,
diseases,
~ generate_forecast(
data = .x,
smooth_data = smooth,
type = .y,
n_days = n_days,
start_date = start_date
)
)
)
names(forecasts_results$forecasts) <- diseases
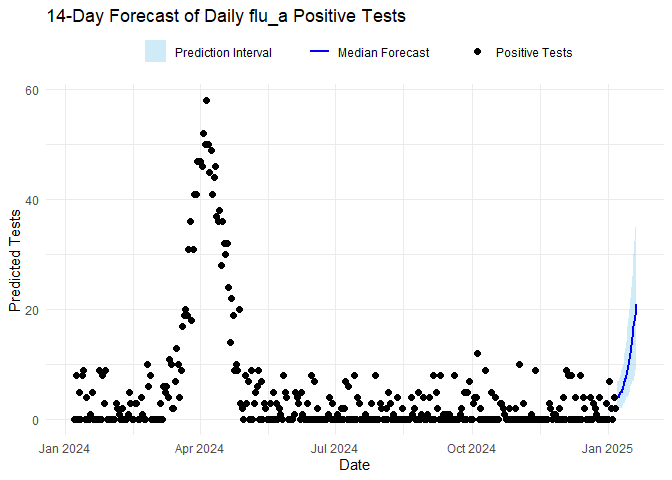
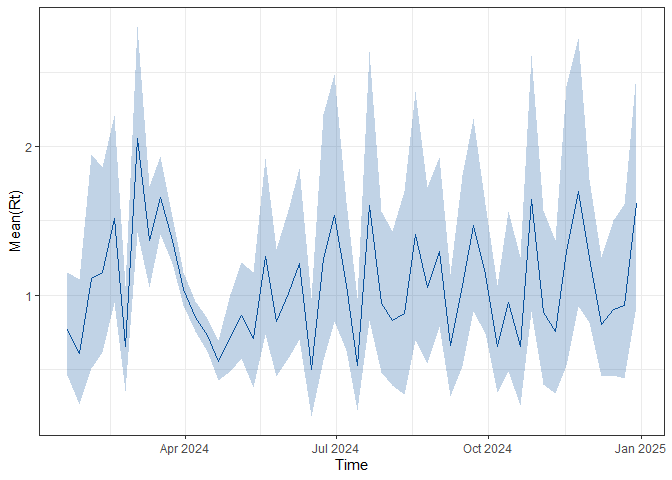
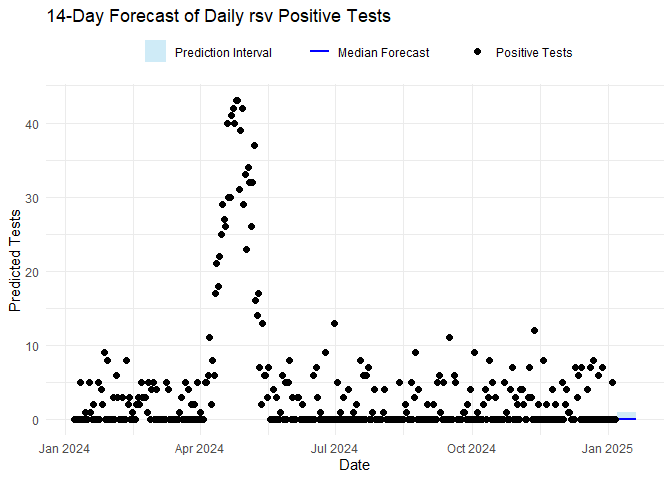
names(forecasts_results$vri_data_list) <- diseasesThe code below plots the forecasts results and the estimated \(R_t\) for each disease. To plot \(R_t\), the code below uses
plot_rt function included in the package.
for (vri in diseases) {
forecast_plot <- ggplot() +
geom_ribbon(
data = forecasts_results$forecasts[[vri]][["forecast_res_quantiles"]],
aes(x = date, ymin = p10, ymax = p90, fill = "Prediction Interval"),
alpha = 0.4
) +
geom_line(
data = forecasts_results$forecasts[[vri]][["forecast_res_quantiles"]],
aes(x = date, y = p50, color = "Median Forecast"),
linewidth = 1
) +
geom_point(
data = forecasts_results$vri_data_list[[vri]],
aes(x = date, y = confirm, shape = "Positive Tests"),
size = 2,
color = "black"
) +
labs(
title = paste0(n_days, "-Day Forecast of Daily ", vri, " Positive Tests"),
x = "Date", y = "Predicted Tests",
fill = "", color = "", shape = ""
) +
scale_fill_manual(values = c("Prediction Interval" = "skyblue")) +
scale_color_manual(values = c("Median Forecast" = "blue")) +
scale_shape_manual(values = c("Positive Tests" = 16)) +
theme_minimal() +
theme(legend.position = "top", legend.direction = "horizontal")
# create Rt plot
rt_plot <- ViroReportR:::plot_rt(forecasts_results$forecasts[[vri]])
print(forecast_plot)
print(rt_plot)
}



Finally, the ViroReportR package can generate an
automated report for the current season across all supported respiratory
viruses (Influenza A, RSV, and SARS-CoV-2) using the
generate_forecast_report() function. This function renders
an HTML report summarizing model outputs and forecasts.
To use it, provide an input file (input_file) containing
the required data with three columns—date, disease_type, and confirm—and
specify an output directory (output_directory) where the
report will be saved.
# rendering forecast report
df <- imap_dfr(vri_data_list, ~ .x %>% mutate(disease_type = .y))
input_file<-"simulated_data.csv"
write.csv(df, input_file, row.names = FALSE)
generate_forecast_report(
input_data_dir = input_file, # input filepath
output_dir = output_directory, # output directory
n_days = 14, # number of days to forecast
validate_window_size = 7, # number of days between each validation window
smooth = FALSE, # logical indicating whether smoothing should be applied in the forecast
)