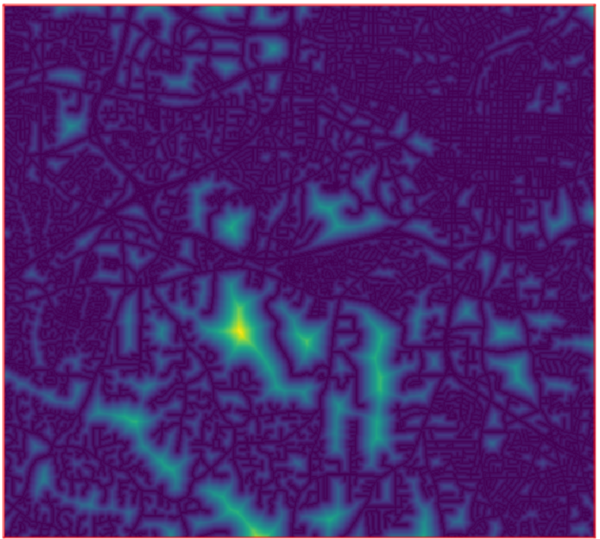
Note: This document is for an older version of GRASS GIS that has been discontinued. You should upgrade, and read the current manual page.

NAME
r.area.createweight - Create a dasymetric weighting layer with Random ForestKEYWORDS
raster, statistics, density, dasymetry, resampleSYNOPSIS
Flags:
- -a
- Use class names for basemap a
- -b
- Use class names for basemap b
- -c
- Keep all covariates in the final model
- -f
- Include detailed results of grid search cross-validation
- --overwrite
- Allow output files to overwrite existing files
- --help
- Print usage summary
- --verbose
- Verbose module output
- --quiet
- Quiet module output
- --ui
- Force launching GUI dialog
Parameters:
- vector=name [required]
- Vector with spatial units
- Polygon vector containing unique ID and response variable in the attribute table
- vector_layer=string [required]
- Layer number or name
- Vector features can have category values in different layers. This number determines which layer to use. When used with direct OGR access this is the layer name.
- Default: 1
- id=name [required]
- Name of the column containing unique ID of spatial units
- Name of attribute column
- response_variable=name [required]
- Name of the column containing response variable
- Format: All values must be >0
- basemap_a=name [required]
- Input raster 1
- E.g. Land cover, Land use, morphological areas...
- basemap_b=name
- Input raster 2 (optional)
- E.g. Land cover, Land use, morphological areas...
- distance_to=name
- Input distance raster (optional)
- Distance to zones of interest
- tile_size=value [required]
- Spatial resolution of output weighting layer
- (in metres)
- output_weight=name [required]
- Output weighting layer name
- Default: weighted_layer
- output_units=name [required]
- Name for output vector gridded spatial units
- Default: gridded_spatial_units
- plot=name [required]
- Name for output plot of model feature importances
- log_file=name [required]
- Name for output file with log of the random forest run
- basemap_a_list=string
- Categories of basemap A to be used
- Format: 1,2,3
- basemap_b_list=string
- Categories of basemap B to be used
- Format: 1,2,3
- n_jobs=integer [required]
- Number of cores to be used for the parallel process
- Default: 1
- kfold=integer
- Number of k-fold cross-validation for grid search parameter optimization
- Format: Must have a value > 2 and < N spatial units
- Default: 5
- param_grid=string
- Python dictionary of customized tunegrid for sklearn RFregressor
Table of contents
DESCRIPTION
r.area.createweight can be used to create a weighting layer for dasymetric mapping using a Random Forest model. Dasymetric mapping is used to redistribute a response variable (e.g. population count) mapped at a coarse spatial unit level (e.g. administrative units), into a raster grid with a finer spatial resolution. Based on a vector layer with the response variable stored in the attribute table and a raster map with categorical values (e.g. land cover), the add-on will produce a raster with the weights to be used in a dasymetric mapping operation that can be performed using v.area.weigh.r.area.createweight also creates a 'gridded' version of the spatial units, whose borders will appear as staircases following the spatial resolution of the weighting layer produced. These two outputs can be used in v.area.weigh module.
A Random Forest (RF) regression model is trained at the level of the vector spatial units. The user must specify a vector layer using the vector parameter. The attribute table connected to this layer must contain a column with a unique identifier (numeric), specified via the id parameter. It must also contain a column with the numeric value to be used as the response variable (e.g. population count), specified via the response_variable parameter.
The user must specify at least one categorical raster map, basemap_a. Optionally, other variables can be added to the Random Forest model: A second categorical raster map can be defined with the parameter basemap_b, and/or a raster with continuous values (e.g. distance to the nearest road, school, hospital, university...) with the parameter distance_to. From these raster maps, the Random Forest's explanatory variables (i.e. covariates, predictors) are calculated. For each of the categorical raster maps (basemaps), the proportion of each class is calculated. For the continuous raster map, the average values are calculated.
Once trained, the Random Forest model predicts weights in a raster grid. The spatial resolution of this output weighted grid should be specified via the tile_size parameter, and its name via the output parameter. The tile_size must be greater than the spatial resolution of basemap_a. It it considered good practice that the tile_size also be greater than the spatial resolution of the other basemap and distance maps. The extent of the output weighted grid is created using the extent of the spatial units, i.e. the vector parameter.
By default, all classes from the raster basemap(s) are taken into account. If the user wants only specific classes to be taken into account within the Random Forest model, it is possible to provide an optional list of these classes with the basemap_a_list parameter (for basemap_a) or the basemap_b_list parameter (for basemap_b, if used).
The out-of-bag error (OOB) of the model is printed in the console and gives an indication about the internal accuracy of the model (cross-validation on training set at the spatial units level). The log of the Random Forest run (including the OOB error) is saved in a file which path and name has to be specified via the log_file parameter. Using the -f flag, the log file will include extended information about the cross-validation for each set of parameters tested in the grid search.
Feature importances in the model are plotted in a graph, which path and name has to be specified via the plot parameter. By default, class values are used. Optionally, class labels can be used as plot labels, using the flags -a and -b, for basemap_a and basemap_b respectively. If these flags are used, the raster should already contain the associated category labels. The module r. category can be used to set category labels for a raster map. If the flag(s) -a and/or -b is(are) selected without existing categories for the corresponding raster map, the class value will be kept.
Parallel processing is supported. The number of cores to be used should be specified via the n_jobs parameter.
The addon is described in more details in a paper [3] with a case study.
NOTES
The module r.mask is used within r.area.createweight. The user should first remove any masks.
It is good practice that the spatial units (vector parameter) are entirely spatially covered by all the input rasters. Any spatial units not sufficiently covered by all the input rasters should first be removed by the user, prior to run r.area.createweight. The add-on, however, will function even if the spatial units are not completely covered by the input rasters. This allows for occasionally missing pixels, or a non-perfect alignment of input rasters with the spatial units (i.e. NULL values in the rasters). In the example below, numerous spatial units are not sufficiently covered, and should be removed prior to running the add-on. It is up to the users to choose an acceptable level of coverage for their analysis, and to ensure the quality of the input data, keeping in mind that the higher the coverage, the better the RF model. The RF model and its' prediction will only be as good as the input data it is given.
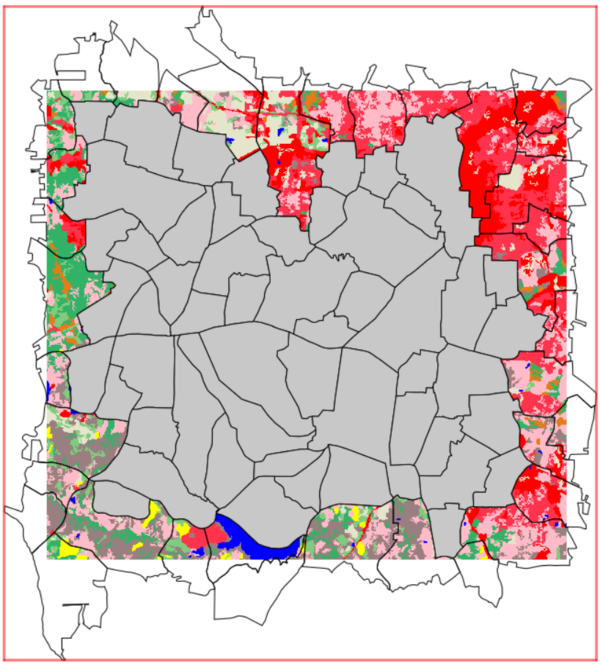
Only the spatial units in grey are completely spatially covered by the basemap and are the ideal selection for the analysis.
If a cell of the weighted grid (output parameter) is not covered by all the input rasters (i.e. if the statistics calculated for that cell are null for at least one of the input rasters), the cell will be given a nodata value.
The module makes a temporary copy of the categorical input rasters (basemap_a and basemap_b) clipped to the area covered by the spatial units. This allows for the extraction of raster categories that exist only within the spatial units. If a user specifies a list of raster categories (basemap_a_list or basemap_b_list), these classes must be present in the area covered by the spatial units. The distance map (distance_to) is not changed, and it is kept in its original format.
The spatial units are rasterised to the extent, cell size and alignment of the output weighted grid, and then are re-vectorised. This results in spatial units whose boundaries will have a 'staircase' appearance, and ensures that each tile of the output weighted grid will be contained in only one spatial unit. This 'gridded' version of the spatial units can be used as an input for dasymetric mapping with v.area.weigh. However, if the original vector contains small (or very narrow) polygons, and the desired tile-size is too large, some polygons can disappear during this process. This will produce an error. The user should reduce the tile size, and/or edit the spatial units vector to merge the smallest spatial units with their neighbouring units (see examples).
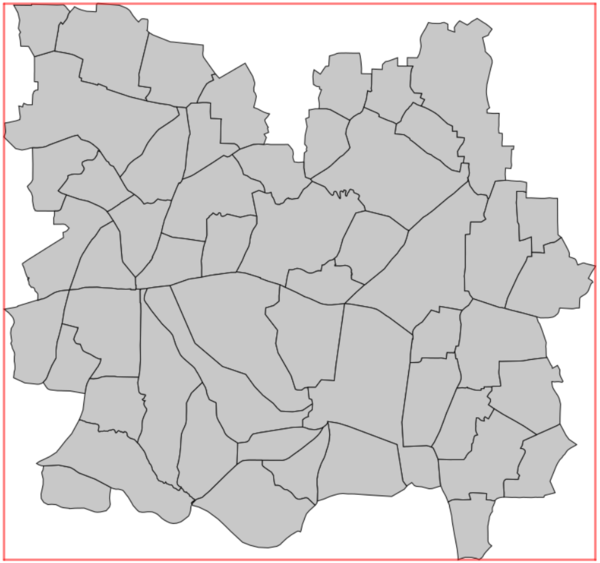 |
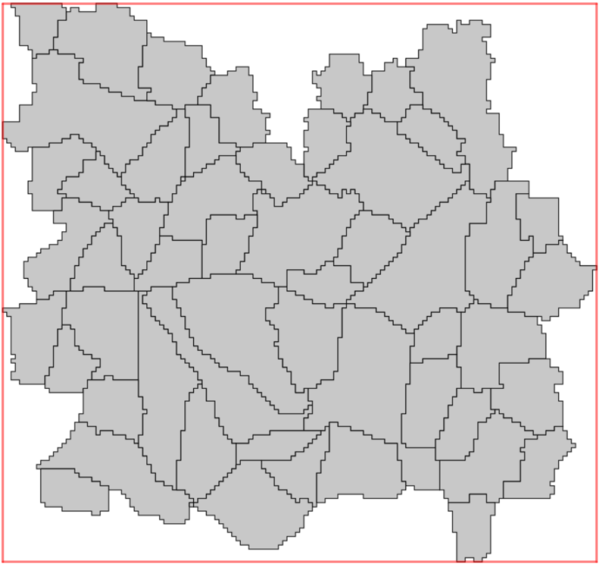
|
|---|---|
| Input spatial units | 'Gridded' spatial units |
The response variable is log-transformed to avoid non-normal distribution and used to train the model. The prediction is then back-transformed and stored in the final weight raster. This approach is similar to the one proposed by [1]. Because of this log transformation (napierian logarithm), it is mandatory to not have zero or negative values in the column containing the response variable (response_variable parameter). It is also expected that the response variable column does not contain NULL values. For the same reason, the model is unable to predict a zero weight value. The implementation of the add-on is designed to set a value of 0 in the weighting layer if the predicted weight is smaller than 0.0000000001 obs./m².
The covariates whose feature importance is bellow 0.5% are, by default, removed from the final model. The -a flag can be used to force keeping all the covariates in the final model.
The parameters of the Random Forest model are tuned using grid search and cross-validation. By default, a 5-fold cross-validation scheme is used but the user can change the number of folds using the kfold parameter. Optionally, the user can provide a Python dictionary with the parameters to be tested via the param_grid parameter. For information about the parameters to be used, please refer to the scikit-learn manual. An example of the format is as follows, these are the parameters tested in the add-on:
"{'oob_score':[True],'bootstrap':[True],\
'max_features':['sqrt',0.1,0.3,0.5,0.7,1],\
'n_estimators':[500,1000]}"
Dependencies
Python 3 is required.GRASS GIS addons
r.area.createweight requires the GRASS GIS addons i.segment.stats, r.zonal.classes, and r.clip to be installed. This can be done using g.extension.Python libraries
r.area.createweight uses the "scikit-learn" machine
learning package (version >= 0.24.1) along with the "pandas"
Python package (version >= 1.0.1). These packages need to be installed
within your GRASS GIS Python environment for
r.area.createweight
to work, i.e. using the GRASS command line terminal for installation.
For Linux users, this package should be available through the
Linux package manager in most distributions (named for example
"python3-scikit-learn").
For MS-Windows users using a 64 bit GRASS, the easiest way is to use the
OSGeo4W
installation method of GRASS, where the Python setuptools can also be
installed. The users can then run 'easy_install pip' to install the pip
package manager. Then, they can download the "Python wheels"
corresponding to each package and install them using the following
command: pip install packagename.whl. Links for
downloading wheels are provided below. The version installed should be
compatible with user's Python version. If GRASS was not installed
using the OSGeo4W method, the pip package manager can be installed by
saving the "get-pip.py" python script provided
here in the folder
containing the GRASS GIS Python environment
(GRASSFOLDER/../etc/python/grass) and executing it with administrator
rights with the following command: python get-pip.py
- Pandas Wheel (mainly relevant to Windows users): https://pypi.python.org/pypi/ pandas
- Scikit-learn Wheel (mainly relevant to Windows users): https://pypi.python. org/pypi/scikit-learn/
- Unofficial Windows compiled binaries from Christoph Gohlke
EXAMPLES
Here we use the GRASS GIS sample North Carolina data set to create a weighting layer of population, using the layers basemap_a = landuse96_28m and vector spatial units = censusblk_swwake. A distance map can be created from the streets_wake layer. Some of the layers must first be prepared as follows.1. Dataset preparation of North Carolina data layers
1.1 Prepare distance layer
A layer containing distance to the nearest street can be created as follows:Set region
g.region raster=landuse96_28m
Rasterize streets
v.to.rast input=streets_wake type=line output=streets_wake_rast \ use=attr attribute_column=F_NODE
Create distance to streets map
r.grow.distance input=streets_wake_rast \ distance=distance_streets
Remove unnecessary files
g.remove -f type=raster name=streets_wake_rast
1.2 Prepare spatial units layer
The vector spatial units layer must be prepared prior to running the module, as there are many small spatial units (polygons) that will get lost during rasterization, and some of the spatial units have a value of zero in the column TOTAL_POP, i.e. the column containing the response variable. In this case, both are solved by merging the smaller spatial units with their neighbouring units, as follows:Set region
g.region vector=censusblk_swwake
Create a vector of points with the centroids of all polygons in the initial census file
v.to.points input=censusblk_swwake type=centroid,face \ output=censusblk_swwake_points
Merge polygons to create larger spatial zones
v.clean input=censusblk_swwake output=censusblk_swwake_merge \ tool=rmarea threshold=1060000
Drop attribute table and connection to layer for merged vector
db.droptable -f table=censusblk_swwake_merge v.db.connect -d map=censusblk_swwake_merge
Add new attribute table to merged vector
v.db.addtable map=censusblk_swwake_merge \ columns="id integer"
Populate attribute table with id and population data (using v.vect.stats to sum up population values)
v.what.vect map=censusblk_swwake_merge column=id \ query_map=censusblk_swwake_points \ query_column=OBJECTID v.vect.stats points=censusblk_swwake_points \ areas=censusblk_swwake_merge method=sum points_column=TOTAL_POP \ count_column=count stats_column=pop
v.extract -r input=censusblk_swwake_merge \ cats='32,552,494,479,483,604,515,553,724,621,700,956,1500,1819,1597, \ 2190,2239,2263,2360,2483,2514,2507,2433,2503,2466,2513,471,472,469, \ 461,448,388,253,204,145,43,33' output=censusblk_swwake_final
Remove unnecessary files
g.remove -f type=vector \ name=censusblk_swwake_points,censusblk_swwake_merge
![[SDF]](r_area_createweight_census_initial.png) |
![[SDF]](r_area_createweight_census_merge.png) |
![[SDF]](r_area_createweight_census_final.png) |
|---|---|---|
| Initial census layer | Census layer after merging | Census layer after removal of non-covered spatial units |
2. Create weighted layer
Generate a weighting layer using a land use mapr.area.createweight vector=censusblk_swwake_final id=cat \ response_variable=pop basemap_a=landuse96_28m tile_size=100 \ output_weight=weighted_layer output_units=gridded_spatial_units \ plot=path/to/filename log_file=path/to/filename n_jobs=4
Generate a weighting layer using only certain classes of the land use map
r.area.createweight vector=censusblk_swwake_final id=cat \ response_variable=pop basemap_a=landuse96_28m tile_size=100 \ output_weight=weighted_layer output_units=gridded_spatial_units \ plot=path/to/filename log_file=path/to/filename \ basemap_a_list=1,2,4 n_jobs=4
Generate a weighting layer using land use map, distance map and associated class names for feature importance plot
r.area.createweight -a vector=censusblk_swwake_final id=cat \ response_variable=pop basemap_a=landuse96_28m \ distance_to=distance_streets tile_size=100 \ output_weight=weighted_layer output_units=gridded_spatial_units \ plot=path/to/filename log_file=path/to/filename n_jobs=4
![[SDF]](r_area_createweight_importances_plot_nonames.png) |
![[SDF]](r_area_createweight_importances_plot_names.png) |
|---|---|
| Feature importances without names | Feature importances with names |
KNOWN ISSUES
On Windows, the installation of Pandas library could be quite difficult to handle (see above for hints).REFERENCES
[1] Stevens, F.R., Gaughan, A.E., Linard, C. and Tatem, A.J., 2015. Disaggregating census data for population mapping using random forests with remotely-sensed and ancillary data. PloS one, 10(2), e0107042. https://doi.org/10. 1371/journal.pone.0107042[2] Grippa, T., Linard, C., Lennert, M., Georganos, S., Mboga, N., Vanhuysse, S., Gadiaga, A., Wolff, E., 2019. Improving urban population distribution models with very-high resolution satellite information. Data, 4(1), 13. https://doi.org/10.3390/ data4010013
[3] Flasse, C., T. Grippa, et S. Fennia. 2021. A TOOL FOR MACHINE LEARNING BASED DASYMETRIC MAPPING APPROACHES IN GRASS GIS. The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences XLVI-4/W2-2021: 55‑62. https://doi.org/10.5194/isprs-archives-XLVI-4-W2-2021-55-2021
ACKNOWLEDGEMENT
This work was funded by the Belgian Federal Science Policy Office (BELSPO) (Research Program for Earth Observation STEREO III), as part of the MAUPP project (contract SR/00/304) and DASYWEIGHT project (contract SR/11/205).SEE ALSO
v.area.weigh (addon), i.segment.stats (addon)AUTHORS
Tais GRIPPA, Safa FENNIA, Charlotte FLASSE - Universite Libre de Bruxelles. ANAGEO Lab.SOURCE CODE
Available at: r.area.createweight source code (history)
Latest change: Saturday Jul 23 07:34:20 2022 in commit: a3c3e0a7c4709e6c4bc16ac9040f95ef6983539c
Main index | Raster index | Topics index | Keywords index | Graphical index | Full index
© 2003-2023 GRASS Development Team, GRASS GIS 7.8.9dev Reference Manual