r.edm.eval
Computes evaluation statistics for a given prediction layer
r.edm.eval [-pb] reference=name prediction=name [,name,...] [csv=name] [figure=name] [buffer=meter] [preval=<0-1>] [plot_dimensions=string] [dpi=integer] [fontsize=float] [n_bins=integer] [presence=integer] [absence=integer] [--overwrite] [--verbose] [--quiet] [--qq] [--ui]
Example:
r.edm.eval reference=name prediction=name
grass.script.run_command("r.edm.eval", reference, prediction, csv=None, figure=None, buffer=None, preval=None, plot_dimensions="6.4,4.8", dpi=100, fontsize=10, n_bins=200, presence=1, absence=0, flags=None, overwrite=None, verbose=None, quiet=None, superquiet=None)
Example:
gs.run_command("r.edm.eval", reference="name", prediction="name")
grass.tools.Tools.r_edm_eval(reference, prediction, csv=None, figure=None, buffer=None, preval=None, plot_dimensions="6.4,4.8", dpi=100, fontsize=10, n_bins=200, presence=1, absence=0, flags=None, overwrite=None, verbose=None, quiet=None, superquiet=None)
Example:
tools = Tools()
tools.r_edm_eval(reference="name", prediction="name")
This grass.tools API is experimental in version 8.5 and expected to be stable in version 8.6.
Parameters
reference=name [required]
Name of raster layer containing references classes
prediction=name [,name,...] [required]
Name of raster layer with predictions
csv=name
Name csv file with evaluation statistics
figure=name
Name of the output file (extension decides format)
Name of the output file. The format is determined by the file extension.
buffer=meter
Restrict absence area
preval=<0-1>
Prevalence of presence points
Allowed values: 0-1
plot_dimensions=string
Plot dimensions (width,height)
Dimensions (width,height) of the figure in inches
Default: 6.4,4.8
dpi=integer
DPI
Resolution of plot in dpi's
Default: 100
fontsize=float
Font size
The basis font size (default = 10)
Default: 10
n_bins=integer
Number of bins in which to divide the modeled distribution scores
Default: 200
presence=integer
Score that represent presence/true
Default: 1
absence=integer
Score that represent absence/false
Default: 0
-p
Print plot to screen
-b
Add presence to background
--overwrite
Allow output files to overwrite existing files
--help
Print usage summary
--verbose
Verbose module output
--quiet
Quiet module output
--qq
Very quiet module output
--ui
Force launching GUI dialog
reference : str, required
Name of raster layer containing references classes
Used as: input, raster, name
prediction : str | list[str], required
Name of raster layer with predictions
Used as: input, raster, name
csv : str, optional
Name csv file with evaluation statistics
Used as: output, file, name
figure : str, optional
Name of the output file (extension decides format)
Name of the output file. The format is determined by the file extension.
Used as: output, file, name
buffer : float, optional
Restrict absence area
Used as: meter
preval : float, optional
Prevalence of presence points
Used as: <0-1>
Allowed values: 0-1
plot_dimensions : str, optional
Plot dimensions (width,height)
Dimensions (width,height) of the figure in inches
Default: 6.4,4.8
dpi : int, optional
DPI
Resolution of plot in dpi's
Default: 100
fontsize : float, optional
Font size
The basis font size (default = 10)
Default: 10
n_bins : int, optional
Number of bins in which to divide the modeled distribution scores
Used as: integer
Default: 200
presence : int, optional
Score that represent presence/true
Used as: integer
Default: 1
absence : int, optional
Score that represent absence/false
Used as: integer
Default: 0
flags : str, optional
Allowed values: p, b
p
Print plot to screen
b
Add presence to background
overwrite : bool, optional
Allow output files to overwrite existing files
Default: None
verbose : bool, optional
Verbose module output
Default: None
quiet : bool, optional
Quiet module output
Default: None
superquiet : bool, optional
Very quiet module output
Default: None
reference : str | np.ndarray, required
Name of raster layer containing references classes
Used as: input, raster, name
prediction : str | list[str], required
Name of raster layer with predictions
Used as: input, raster, name
csv : str, optional
Name csv file with evaluation statistics
Used as: output, file, name
figure : str, optional
Name of the output file (extension decides format)
Name of the output file. The format is determined by the file extension.
Used as: output, file, name
buffer : float, optional
Restrict absence area
Used as: meter
preval : float, optional
Prevalence of presence points
Used as: <0-1>
Allowed values: 0-1
plot_dimensions : str, optional
Plot dimensions (width,height)
Dimensions (width,height) of the figure in inches
Default: 6.4,4.8
dpi : int, optional
DPI
Resolution of plot in dpi's
Default: 100
fontsize : float, optional
Font size
The basis font size (default = 10)
Default: 10
n_bins : int, optional
Number of bins in which to divide the modeled distribution scores
Used as: integer
Default: 200
presence : int, optional
Score that represent presence/true
Used as: integer
Default: 1
absence : int, optional
Score that represent absence/false
Used as: integer
Default: 0
flags : str, optional
Allowed values: p, b
p
Print plot to screen
b
Add presence to background
overwrite : bool, optional
Allow output files to overwrite existing files
Default: None
verbose : bool, optional
Verbose module output
Default: None
quiet : bool, optional
Quiet module output
Default: None
superquiet : bool, optional
Very quiet module output
Default: None
Returns:
result : grass.tools.support.ToolResult | None
If the tool produces text as standard output, a ToolResult object will be returned. Otherwise, None will be returned.
Raises:
grass.tools.ToolError: When the tool ended with an error.
DESCRIPTION
The r.edm.eval addon returns a few evaluation statistics, including the area under the curve (AUC), the maximum true skill statistic (TSS), and the maximum kappa. For all three statistics, the corresponding probability threshold values are also given. Optionally, it draws the receiver operating characteristic (ROC) curve.
The addon takes as input a raster layer with observations(observations) and one or more predictions layers. The observations layer should be encoded with 1 (presence/cases) and 0 (absences/controls). The (predictions layer gives the predicted values (e.g., the outcome of a model). This typically are probability or suitability scores between 0 and 1, but it will work on other scales too. With two or more predictions layers, statistics are calculated for each prediction layer separately. This allows one to compare predictions of different models.
By default, the prediction layer is divided in 200 bins (the user can change this number). For each bin, the module computes the cumulative true and false positives (TP, FP), true and false negatives (TN, FN), the true and false positive rate (TPR, FPR), the true negative rate (TNR), Cohen's kappa and the true skill statistic.
TPR = TP / (TP + FN)
FPR = FP / (FP + TN)
TNR = TN / (TN + FP)
TSS = TPR - FPR
Kappa = 2 * (TP * TN - FN * FP) / ((TP + FP) * (FP + TN) + (TP + FN) * (FN + TN)))
These values can be saved to a CSV file, together with the corresponding upper and lower boundaries of the bins values. These can be used to calculate additional statistics.
With the -b flag, the module will use the fraction of points that is predicted to be presence/true instead of the false positive rate to create the ROC curve and calculate the AUC. Use this when the predictions are created using a model that works with background points instead of (pseudo-)absence points (like Maxent).
A problem with the AUC is that it varies with the spatial extent used to select background points (Lobo et al. 2008, Jiménez-Valverde 2012). Generally, the larger that extent, the higher the AUC value. Therefore, AUC values are generally biased and cannot be directly compared. An admittedly simple way to evaluate this issue of changing AUC values with changing extents is to only use (pseudo-)absence or background points within certain distances of the presence points (raster cells). To do so, the user can use the buffer parameter to limit the absence/background points that are used to calculate the various statistics to those within a certain distance from the presence locations/areas.
The user can set the required ratio of presence and total points to be used in the evaluation (preval; value between 0 an 1). This allows the user to test how sensitive the evaluation statistics are for the prevalence of presence points.
NOTES
The module expects an reference raster layer, encoded with 1 (presence/cases) and 0 (absences/controls). Optionally, the user can define other values using the absence and presence parameters. If the layer contains more than 2 unique values, or if the layer is not of the type CELL (integer), the module will terminate with a warning.
The selection of the best evaluation statistic depends on your data and what you want to test. This module provides a few only, but the user can calculate his/her own using the table in the CSV file. See the References list for a few useful references in that respect.
This module depends on the Python Pandas library.
EXAMPLES
Run this example in the "landsat" mapset of the North Carolina sample data set location. First, create a binary map with herbaceous land cover (1) and other (0). Next, create a raster layer with training pixels using r.learn.train and r.learn.predict from the r.learn.ml2 addon. Note, r.learn.train can compute its own accuracy measures as well.
g.region raster=landclass96
r.mapcalc "herbaceous = if(landclass96 == 3, 1, 0)"
r.random input=herbaceous npoints=1000 raster=training_pixels seed=10
Then we can use these training pixels to perform a classification on the more recently obtained landsat 7 image using r.learn.train.
# train a random forest regression model using r.learn.train
r.learn.train group=lsat7_2000 training_map=training_pixels \
model_name=RandomForestRegressor n_estimators=500 \
save_model=rf_model.gz cv=2
# perform prediction using r.learn.predict
r.learn.predict group=lsat7_2000 load_model=rf_model.gz \
output=rf_regressor
Now we can see how well the model performed using the r.edm.eval.
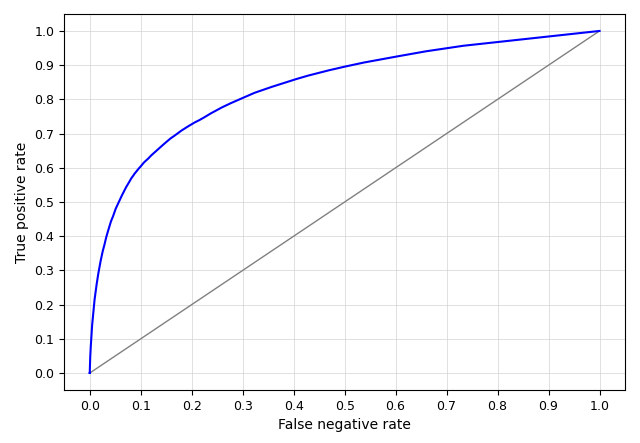
r.edm.eval -p reference=herbaceous prediction=rf_regressor
The accuracy measures are printed on screen.
AUC = 0.8362
maximum TSS = 0.5289
maximum kappa = 0.4438
treshold maximum TSS = 0.1118
treshold maximum kappa = 0.2936
treshold minimum distance to (0,1) = 0.0979
And the accompanying ROC curve is shown below (the figure can optionally be saved to file).
Let's try how well a logistic regression performs, and compare this with the previous results.
# train a logistic regression model using r.learn.train
r.learn.train group=lsat7_2000 training_map=training_pixels \
model_name=LogisticRegression save_model=lr_model.gz cv=2
# perform prediction using r.learn.predict -p
r.learn.predict group=lsat7_2000 load_model=lr_model.gz \
output=lr_regressor
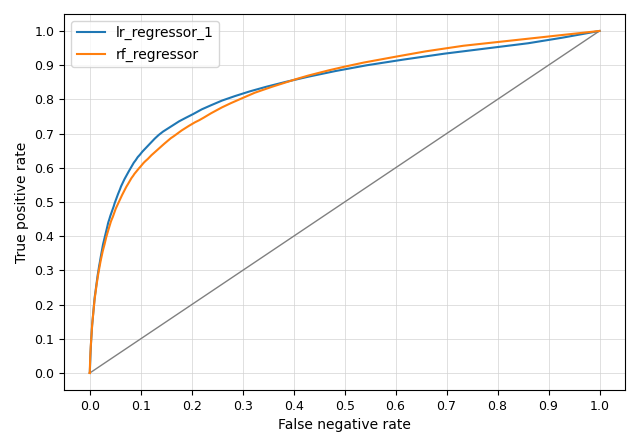
r.edm.eval -p reference=herbaceous prediction=rf_regressor,lr_regressor_1
The accuracy measures are printed on screen.
AUC = 0.8362
maximum TSS = 0.5289
maximum kappa = 0.4438
treshold maximum TSS = 0.1118
treshold maximum kappa = 0.2936
treshold minimum distance to (0,1) = 0.0979
--- performance lr_regressor_1 ---
AUC = 0.8384
maximum TSS = 0.5621
maximum kappa = 0.4669
treshold maximum TSS = 0.1047
treshold maximum kappa = 0.1995
treshold minimum distance to (0,1) = 0.0848
And the accompanying ROC curve is shown below (the figure can optionally be saved to file).
REFERENCES
Jiménez-Valverde, A. 2012. Insights into the area under the receiver operating characteristic curve (AUC) as a discrimination measure in species distribution modelling. Global Ecology and Biogeography 21: 498-507
Lobo, J. M., Jiménez-Valverde, A., & Real, R. 2008. AUC: a misleading measure of the performance of predictive distribution models. Global Ecology and Biogeography 17: 145-151.
Hijmans, R. J. 2012. Cross-validation of species distribution models: removing spatial sorting bias and calibration with a null model. Ecology 93: 679-688.
Allouche, O., Tsoar, A., & Kadmon, R. 2006. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). Journal of Applied Ecology 43: 1223-1232.
McPherson, J. M., Jetz, W., & Rogers, D. J. 2004. The effects of species' range sizes on the accuracy of distribution models: ecological phenomenon or statistical artefact? Journal of Applied Ecology 41: 811-823.
SEE ALSO
r.confusionmatrix, r.learn.ml2
AUTHOR
Paulo van Breugel
SOURCE CODE
Available at: r.edm.eval source code
(history)
Latest change: Thursday Mar 20 21:36:57 2025 in commit 7286ecf